rgdal/sp packages were obsolete and needed to be replaced with package sf, but I am not yet familiar enough with sf so here it is with rgdal/sp:
library(rgdal)
setwd("~/Desktop") #For code simplicity I am explicitely setting a working directory in which every file will be.
nsb <- NSBcompanion::nsbConnect("guest","arm_aber_sexy")
exp379 <- dbGetQuery(nsb,"SELECT hole_id, longitude, latitude FROM neptune_hole_summary WHERE leg=379 and hole='A';")
dbDisconnect(nsb)
exp379sp <- SpatialPointsDataFrame(exp379[,-1],data=exp379, proj4string=CRS("+proj=longlat"))
writeOGR(exp379sp,".","exp379",driver="ESRI Shapefile") #"." because we save it in the current folderdownload.file("https://www.earthbyte.org/webdav/ftp/Data_Collections/Matthews_etal_2016_Global_Plate_Model_GPC.zip","Matthews_etal_2016.zip") #download
unzip("Matthews_etal_2016.zip") #unzipsystem("gplates assign-plate-ids -p Matthews_etal_2016_Global_Plate_Model_GPC/Global_EarthByte_Mesozoic-Cenozoic_plate_boundaries_Matthews_etal.gpml\\
-l exp379.shp -t 0 -s shapefile")
# -p is the plate boundary file that will be used to partition the sites
# -l is the feature we want to partition
# -t is the time at which the features are taken
# -s is the format we want to save the result as
# Just to check it worked, read in the file and check it has plate IDs
e3 <- readOGR(".","exp379")
e3$PLATEID1 #Should show 802 for both sites.
rm(e3)system("gplates reconstruct -t 90 -l exp379.shp\\
-r Matthews_etal_2016_Global_Plate_Model_GPC/Global_EB_250-0Ma_GK07_Matthews_etal.rot\\
-o exp379_cretaceous -e shapefile")
system("gplates reconstruct -t 90 -l Matthews_etal_2016_Global_Plate_Model_GPC/StaticGeometries/Coastlines/Global_coastlines_2015_v1_low_res.shp \\
-r Matthews_etal_2016_Global_Plate_Model_GPC/Global_EB_250-0Ma_GK07_Matthews_etal.rot\\
-o coast_cretaceous -e shapefile")exp379cret <- readOGR(".","exp379_cretaceous")
coast <- readOGR(".","coast_cretaceous")
dest_projection <- CRS("+proj=laea +lat_0=-90 +lon_0=0")
polar <- spTransform(coast,dest_projection)
polar_379 <- spTransform(exp379cret,dest_projection)
polar_poly <- as(polar,"SpatialPolygons") #Here the coastlines are indeed lines, but we want them as polygons so that we can color-fill them.
limitANT <- Lines(list(Line(cbind(-180:180,-45))),ID="a")
limitANT <- SpatialLines(list(limitANT),proj4string=CRS("+proj=longlat"))
limitANT <- spTransform(limitANT,dest_projection)
png("turonian.png",h=400,w=400)
par(mar=c(0,0,0,0))
plot(limitANT,col="white")
plot(polar_poly,col="grey80",border="grey80",add=TRUE)
plot(polar_379,col="red",pch=19,add=TRUE,cex=0.5)
box(lwd=3)
dev.off()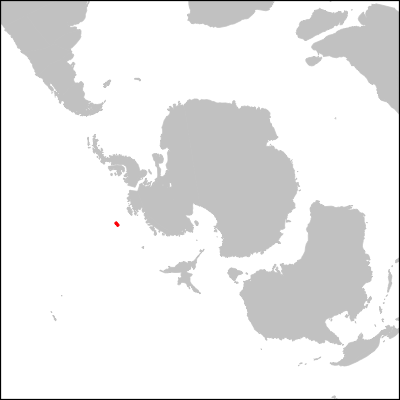
I know, it's a bit underwhelming since the Antarctic plate didn't move that much in the last 100Myr, but you get the gesture :)
